New 3D interactive software unravels ‘fabric of life’
Primary page content
Dynamic new interactive technology which visualises the 3D structures inside DNA has been launched by a team of computational artists, game developers and scientists, working together to help the public better understand the cause of diseases.
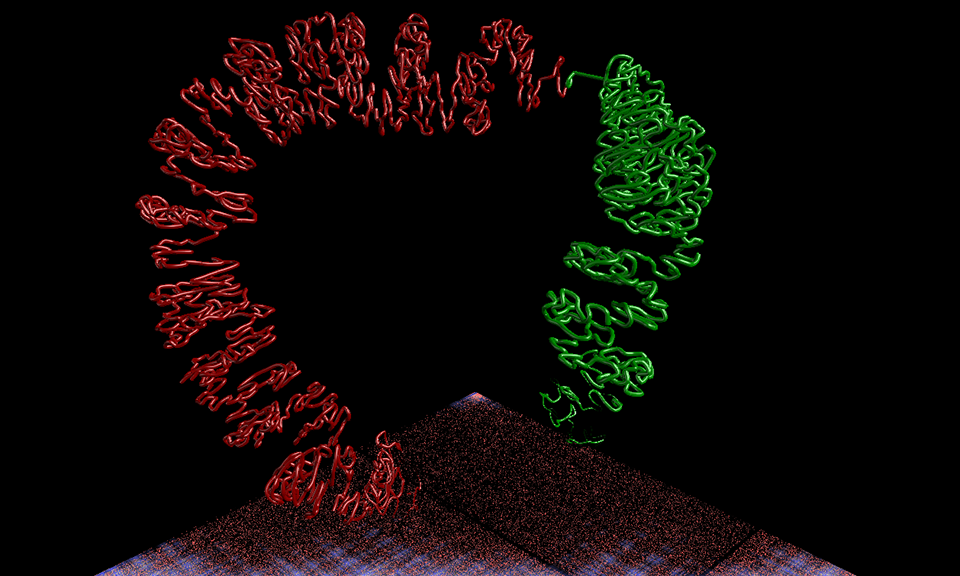
Chromatin mitosis
CSynth is a software platform created by researchers at Goldsmiths, University of London and Oxford University. Described by its designers as ‘bio-visualisation made interactive’, it shows how cell machinery physically interacts with a structure as complex and compact as the genome.
Viewers can watch and explore the 3D models on a screen, or use a Virtual Reality headset to immerse themselves in genetic material and manipulate it themselves.
Traditionally, scientists have only been able to visualise and understand the genome - the complete set of genetic material present in a cell - in 2D presentations, on a screen or through graphs or histograms.
But as researchers gather more data about how cells work it is clear that a 3D structure is extremely important for gene regulation and how cells differentiate. For example, a white blood cell looks and behaves differently to a red blood cell even though its genome is exactly the same.
Subtle differences in the way the genome is folded can impact on whether genes can be switched on and off at particular times, which then dictates what a cell can do. Changes in the way chromatin is folded can cause rare blood diseases, for example, because it impacts on how genetic code is read by a cell.
Understanding this process is vital for seeking the cause of diseases such as diabetes or anaemia, and for the development of treatments for them.
Thanks to advances in genetic techniques, researchers are able to harness more information than ever before from biological data provided by patients and volunteers.
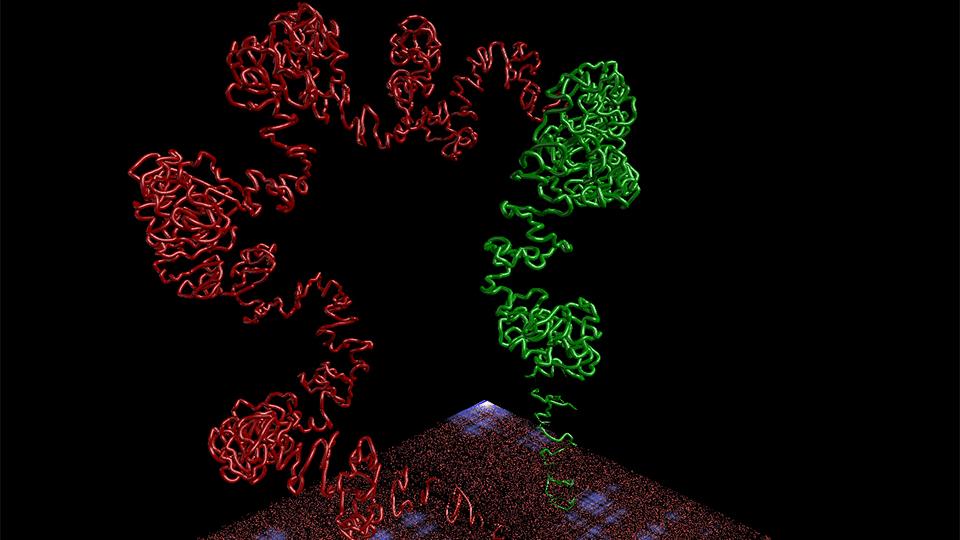
(Chromatin interphase)
The CSynth software then integrates data from genome sequencing and computer modelling and presents it in an attractive and engaging way, using computer game technology.
The team have launched a complete software package that will also allow the import of public data, and help both the public and medical researchers gain a better understanding of how the genome is folded in a cell, and the complex mechanisms involved.
Professor Frederic Fol Leymarie and Professor William Latham from the Department of Computing at Goldsmiths are the computer artists and software designers behind CSynth, working with Steve Taylor, Head of Analysis, Visualisation and Informatics at the WIMM Centre of Computational Biology, and Professor Jim Hughes at the MRC Weatherall Institute of Molecular Medicine, University of Oxford. They are joined by Professor Stephen Todd, lead software architect at London Geometry Ltd and Visiting Professor in Computing at Goldsmiths, and Peter Todd, senior developer, London Geometry Ltd.
Steve Taylor said: “We have made a web-based interface where any researcher can load in the data from their experiments. Previously the software had to be installed and all the parameters were adjusted in text files by us behind the scenes. Now you can upload or drag and drop the data into a web page, and it will build a model allowing investigators to really get a handle on their data. You also get a fantastic user interface to interact with the model and overlay other data, such as genes and enhancers. We get asked a lot about making CSynth available for teaching and and now we can do this easily.”
Professor Fol Leymarie said: “Our body is made of trillions of cells, each one containing chromatin tightly folded. This very long molecular strand is not static, but rather keeps moving, vibrating, unfolding and refolding locally, more like a molecular dance.
“Furthermore, it keeps interacting with other molecular structures present in the cell and with itself. It is this dynamic nature that CSynth makes visible and interactive, so that a user - a researcher, student or even a curious member of the public - can load different data sequences, try out various parameters, compare various situations, to eventually get a much better, intuitive understanding, which we hope may help lead to new discoveries.”
Visit www.csynth.org or read the paper CSynth: A Dynamic Modelling and Visualisation Tool for 3D Chromatin Structure on the open access bioRxiv platform for more information.